health-care-markets
Methods and thoughts on defining geographic markets for health care services, i.e., a guided tour of a particularly complex rabbit hole.
construct-dartmouth-geography-data.R
johngraves Tue Jan 29 21:33:38 2019
####################################################
####################################################
####################################################
# Create HRR and HSA Boundary and Data Files
####################################################
####################################################
####################################################
# The objective of this document is to construct the HRR, HSA, and PCSA
# boundary and data files from the underlying ZCTA data.
# The reason we do this over using the boundary files constructed by Dartmouth
# is that those files lack sufficient information to obtain contiguous
# market area data.
# Also, this file serves as a blueprint more generally for obtaining
# polygon data frames and contiguous areas data for any clustering of
# a smaller geographic unit (e.g., counties to markets).
suppressWarnings(suppressMessages(source(here::here("/R/manifest.R"))))
source(here("R/move-ak-hi.R"))
source(here("R/get-geographic-info.R"))
source(here("R/map-theme.R"))
source(here("R/shared-objects.R"))
source(here("R/get-contiguous-areas.R"))
# Get information on PCSA state, etc. (this is merged on later)
pcsa_info <- foreign::read.dbf(here("public-data/shape-files/dartmouth-hrr-hsa-pcsa/ct_pcsav31.dbf")) %>%
janitor::clean_names() %>%
select(pcsa,pcsa_st, pcsa_l) %>%
mutate_at(vars(pcsa,pcsa_st,pcsa_l),funs(paste0)) %>%
unique()
# ZCTA to PCSA Crosswalk (merges on PCSA state from above)
zcta_to_pcsa <- foreign::read.dbf(here("public-data/shape-files/dartmouth-hrr-hsa-pcsa/zip5_pcsav31.dbf")) %>%
janitor::clean_names() %>%
mutate(pcsa = paste0(pcsa)) %>%
left_join(pcsa_info,"pcsa")
# ZCTA to HRR and HSA Crosswalk
zcta_to_hrr_hsa <- read_csv(here("public-data/shape-files/nber-hrr-hsa-pcsa/ziphsahrr2014.csv")) %>%
janitor::clean_names() %>%
rename(zip5 = zipcode )
## Parsed with column specification:
## cols(
## zipcode = col_character(),
## hsanum = col_double(),
## hsacity = col_character(),
## hsastate = col_character(),
## hrrnum = col_double(),
## hrrcity = col_character(),
## hrrstate = col_character(),
## year = col_double()
## )
zcta_to_state <- read_csv(here("public-data/zcta-to-fips-county/zcta-to-fips-county.csv")) %>%
filter(row_number() !=1) %>%
rename(zip5 = zcta5) %>%
select(zip5,state = stab) %>%
unique() %>%
group_by(zip5) %>%
mutate(n=paste0("state_",str_pad(row_number(), width=2,pad="0"))) %>%
filter(zip5!="99999") %>%
spread(n,state)
## Parsed with column specification:
## cols(
## state = col_character(),
## zcta5 = col_character(),
## county = col_character(),
## stab = col_character(),
## cntyname = col_character(),
## zipname = col_character(),
## intptlon = col_character(),
## intptlat = col_character(),
## pop10 = col_character(),
## afact = col_character()
## )
# Load the ZCTA Map Shape and merge in HRR, HSA, and PCSA Information
zcta_map_shape <- sf::read_sf(here("public-data/shape-files/zcta-2017/tl_2017_us_zcta510/tl_2017_us_zcta510.shp")) %>%
sf::st_transform(crs ="+proj=aea +lat_1=29.5 +lat_2=45.5 +lat_0=37.5 +lon_0=-96") %>%
janitor::clean_names() %>%
mutate(zip5 = zcta5ce10) %>%
left_join(zcta_to_state,"zip5") %>%
filter(state_01 %in% states) %>%
left_join(zcta_to_pcsa,"zip5") %>%
left_join(zcta_to_hrr_hsa,"zip5")
## Warning: Column `zip5` joining character vector and factor, coercing into
## character vector
df_hrr <-
zcta_to_hrr_hsa %>%
janitor::clean_names() %>%
select(contains("hrr")) %>%
unique()
df_hsa <-
zcta_to_hrr_hsa %>%
janitor::clean_names() %>%
select(contains("hsa")) %>%
unique()
df_pcsa <-
zcta_to_pcsa %>%
janitor::clean_names() %>%
select(contains("pcsa")) %>%
unique()
####
# Construct shapefiles based on the intersection with HRR, HSA, and PCSA
####
sf_state <- sf::read_sf(here("output/tidy-mapping-files/state/01_state-shape-file.shp")) %>%
sf::st_transform(crs ="+proj=aea +lat_1=29.5 +lat_2=45.5 +lat_0=37.5 +lon_0=-96") %>%
st_simplify(dTolerance = 500) %>%
filter(stusps %in% c("TX","HI","AK")) %>%
move_ak_hi(state = stusps)
sf_hrr <- zcta_map_shape %>%
#filter(hrrstate %in% c("TX","HI","AK")) %>%
group_by(hrrnum) %>%
summarise() %>%
mutate(test = as.factor(sample(1:10,nrow(.),replace=TRUE))) %>%
st_simplify(dTolerance = 500) %>%
left_join(df_hrr, "hrrnum") %>%
move_ak_hi(state = hrrstate)
sf_hrr_final <- sf_hrr %>%
left_join(get_contiguous(shp = sf_hrr, id = hrrnum) %>% mutate(hrrnum = as.numeric(paste0(hrrnum))), "hrrnum")
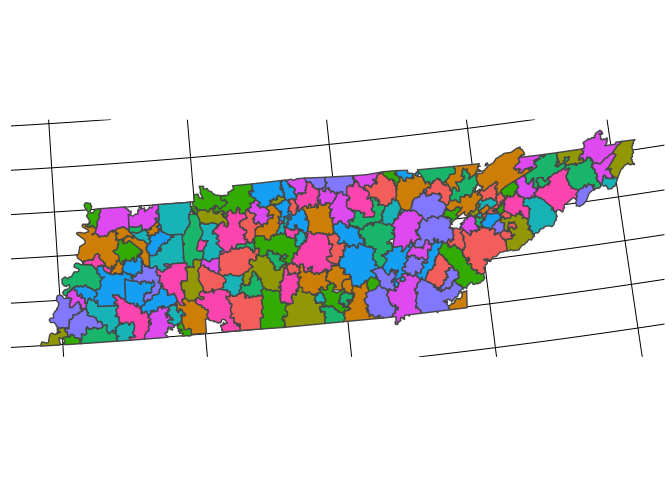
sf_hrr_final %>%
filter(hrrstate=="TN") %>%
ggplot() +
geom_sf(aes(fill = test)) +
remove_all_axes +
theme(legend.position = "none")
sf_hrr_final %>%
sf::write_sf(here("output/tidy-mapping-files/hrr/01_hrr-shape-file.shp"))
sf_hsa <- zcta_map_shape %>%
group_by(hsanum) %>%
summarise() %>%
mutate(test = as.factor(sample(1:10,nrow(.),replace=TRUE))) %>%
st_simplify(dTolerance = 500) %>%
left_join(df_hsa, "hsanum") %>%
move_ak_hi(state = hsastate)
sf_hsa_final <-
sf_hsa %>%
left_join(get_contiguous(shp = sf_hsa, id = hsanum) %>% mutate(hsanum = as.numeric(paste0(hsanum))), "hsanum")
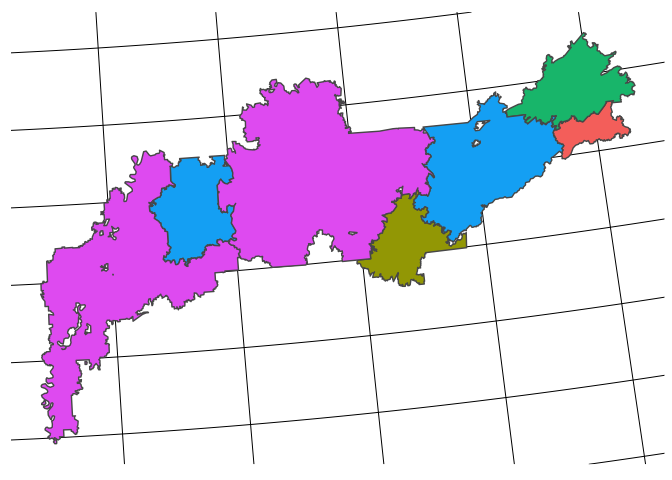
sf_hsa_final %>%
filter(hsastate=="TN") %>%
ggplot() +
geom_sf(aes(fill = test)) +
remove_all_axes +
theme(legend.position = "none")
sf_hsa_final %>%
sf::write_sf(here("output/tidy-mapping-files/hsa/01_hsa-shape-file.shp"))
sf_pcsa <- zcta_map_shape %>%
#filter(hrrstate %in% c("TN","HI","AK")) %>%
group_by(pcsa) %>%
summarise() %>%
mutate(test = as.factor(sample(1:10,nrow(.),replace=TRUE))) %>%
st_simplify(dTolerance = 500) %>%
left_join(df_pcsa, "pcsa") %>%
move_ak_hi(state = pcsa_st)
sf_pcsa_final <-
sf_pcsa %>%
left_join(get_contiguous(shp = sf_pcsa, id = pcsa), "pcsa")
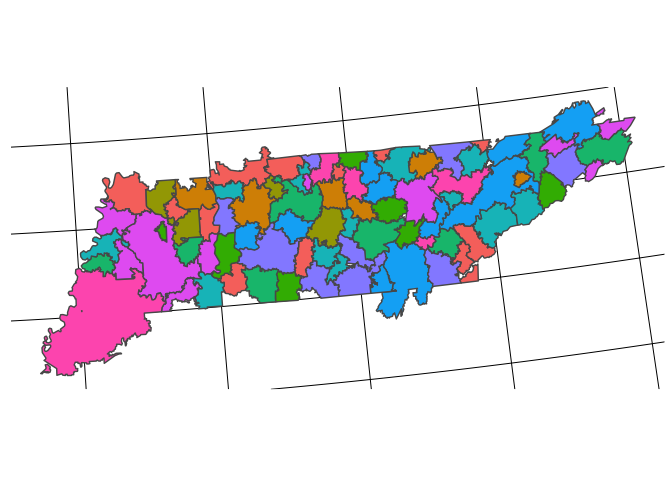
sf_pcsa_final %>%
sf::write_sf(here("output/tidy-mapping-files/pcsa/01_pcsa-shape-file.shp"))
sf_pcsa_final %>%
filter(pcsa_st=="TN") %>%
ggplot() +
geom_sf(aes(fill = test)) +
remove_all_axes +
theme(legend.position = "none")